Appendix C. |
|
|
This appendix is intended for SeqWeb users who also use SeqLab (the X Windows-based interface) or the command-line interface to the Wisconsin Package, and may want to move sequence files between them. |
If you used the GCG Wisconsin Package via SeqLab or the command-line interface in the past, you know that to do so you needed an account on the UNIX or OpenVMS computer running the Package. An account is accessed through a username and password and consists of a directory and the files and subdirectories it may contain.
Your SeqWeb account is separate from your UNIX or OpenVMS account to the Wisconsin Package. You will need to log in to SeqWeb separately, and your SeqWeb directory is separate from any other directories you may have on your UNIX system. The only way to access your SeqWeb directory is by using SeqWeb with a web browser.
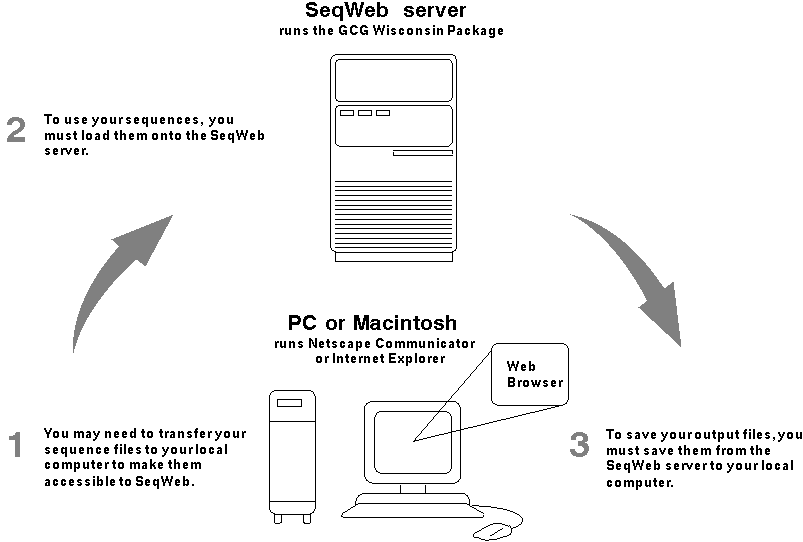
If you previously used the command-line or SeqLab interfaces to the Wisconsin Package on UNIX or OpenVMS, you may have sequences in a GCG format that you want to use in SeqWeb. To do so, you may need to transfer those sequence files, for example via FTP, from the UNIX or OpenVMS computer to SeqWeb (see figure).

Note that if you have network access from your PC or Macintosh to your UNIX or OpenVMS account, you can directly access the files from your web browser and load them into SeqWeb. Alternatively, you may be able to copy and paste your sequences into SeqWeb.
In SeqLab or the command-line Wisconsin Package, you sometimes need to run two or more Wisconsin Package programs to perform a particular type of analysis. In SeqWeb, some of the frequently used multi-program combinations have been seamlessly linked together to look like a single program.
| This SeqWeb Program ... | Runs these Programs, in this Order |
| Compare | Compare, DotPlot |
| GrowTree | PileUp, Distances, GrowTree |
| HmmerBuild | HmmerBuild, HmmerCalibrate (as a Selectable Option) |
| MFold | MFold, PlotFold |
| PeptideStructure | PeptideStructure, PlotStructure |
| Pretty | PileUp, Pretty |
| ProfileSearch | PileUp, ProfileSegments, ProfileMake, ProfileSearch |
| StemLoop | StemLoop, DotPlot |
Note: Although the related programs MEME and MotifSearch are not linked as described above, SeqWeb allows you to run MotifSearch directly from the MEME results page. Likewise, HmmerSearch and HmmerAlign can be run from the HmmerBuild results page.
SeqWeb accepts most GCG sequence file formats, including:
Note: SeqWeb reformats MSF and RSF files and creates single sequence files out of the multiple sequences they contain. Note that SeqWeb does not accept list file format.
SeqWeb also accepts the following non-GCG sequence formats:
What is a list file?
A list file contains a list of sequence names and their locations. No sequence residues are recorded in the file. List files are created by SeqLab, as well as by a number of database searching programs in the Wisconsin Package.
How can I use list files?
With SeqWeb
SeqWeb does not allow you to load list files directly, nor does it accept list files as sequence input. If you have a list file from which you would like to load sequences into SeqWeb, you can do so by reformatting the list file to an RSF file.
- Open a UNIX window and locate the list file.
- From the command line, type % reformat -rsf @list_filename
Substitute the name of your list file for list_filename.
The file is reformatted to an RSF file. When you load the RSF file into SeqWeb, each sequence it contains is extracted and loaded separately.
With SeqLab and Command-Line Interface
List files are a central concept to using SeqLab, where they are used to organize a variety of sequences (from databases, MSF, RSF, and even other list files) on a project-by-project basis.
In addition, some programs in SeqLab and the command-line Wisconsin Package accept list files as input.
A number of SeqWeb programs create list files as output. To use these files with SeqLab or the command-line Wisconsin Package, save them as "Text" as explained in the Saving Results section of the Results Manager chapter.
What is an MSF file?
Multiple Sequence Format (MSF) files contain multiple sequences aligned together. The programs that create MSF files as their output are identified in the File Formats Created by SeqWeb Programs section below.
How can I use MSF files?
With SeqWeb
When you load an MSF file into SeqWeb, each individual sequence within the file is extracted and loaded separately.
With SeqLab and Command-Line Interface
A number of SeqWeb programs create MSF files as output. To use SeqWeb's MSF files with SeqLab or the command-line Wisconsin Package, save them as "Text" as explained in the Saving Results section of the Results Manager chapter.
What are RSF files?
A Rich Sequence Format (RSF) file contains one or more sequences, as well as richly annotated feature information for each sequence. The sequences may or may not be related.
How can I use RSF files?
With SeqWeb
When you load an RSF file into SeqWeb, each individual sequence within the file is extracted and loaded separately.
With SeqLab and Command-Line
RSF files are very useful in SeqLab, where the sequences and their features can be graphically displayed and manipulated. SeqLab's Editor mode lets you create RSF files containing one or more sequences.
SeqWeb cannot create RSF files as output; thus, there is no way to move RSF files from SeqWeb to SeqLab or the command-line Wisconsin Package.
Most SeqWeb programs create text files containing reports of the analysis performed. However, some programs create list, MSF, and GCG-formatted sequence files, as indicated in the following table.
| Program | List File | MSF File | Sequence File |
| BackTranslate | X | ||
| BLAST | X | ||
| FastA | X | ||
| GrowTree | X | ||
| LookUp | X | ||
| MotifSearch | X | ||
| PileUp | X | ||
| ProfileSearch | X | ||
| Reverse | X | ||
| SSearch | X | ||
| StringSearch | X | ||
| Translate | X |