- Background
- Your Task: Run a gel
This chapter introduces you to Vector NTI’s Gel Display windows, powerful tools for running and analyzing gel electrophoresis. Vector NTI predicts the results of actual gel electrophoresis experiments and displays them in text and graphics form.
Your Task: Run a gel
Your task will be to open a new Gel Display window, add samples and gel markers to it, run the gel, and explore NTI’s powerful graphical capabilities for manipulating gels.
Follow the steps in the order shown. Figures show what your screen should look like at various points along the way.
1. Launch Vector NTI
Launch Vector NTI by double-clicking its icon in the program group or folder in which you installed Vector NTI.
2. Create a new Gel and open a Display window for it
Switch to the Vector NTI workspace window and click the New Gel button (  ) on the main toolbar to create a new Gel and to open a Display window for it. A dialog box appears, allowing you to set up the new Gel Display window as desired. You may describe your own gel using the following parameters:
) on the main toolbar to create a new Gel and to open a Display window for it. A dialog box appears, allowing you to set up the new Gel Display window as desired. You may describe your own gel using the following parameters:
gel type (agarose, polyacrylamide, ...)
gel concentration
gel dimensions
voltage
electric field type (steady, pulse)
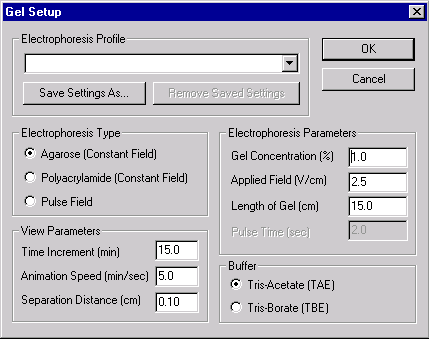
Note that you can set the type and parameters of electrophoresis, select buffers and display parameters, or choose from a list of saved electrophoresis settings. Select the “Example of Agarose Gel” from the Electrophoresis Profile list and press the OK button. Vector NTI creates your new gel and opens it in a Gel Display window.
3. Create a sample and add it to the gel
The Display window you have just created shows an empty gel ready to receive samples.
Let’s create a new sample and add it to the gel. The sample will consist of the SV40 molecule cut by the HaeIII and HindIII restriction endonucleases.
Click the Create Gel Sample button (  ) on the Window toolbar. A dialog box appears, allowing you to create the sample.
) on the Window toolbar. A dialog box appears, allowing you to create the sample.
In the Molecules box, select SV40. In the Enzymes box, use the scroll bar to move up and down through the list of enzymes. Select the HaeIII and HindIII RENs. As you select the molecule and RENs, their names appear in the Selected for Gel Sample column on the right side of the dialog box.
In the Sample Name box near the bottom of the dialog, enter “SAMPLE1”.
In the Description box, enter “SV40 cut by HaeIII and HindIII”.
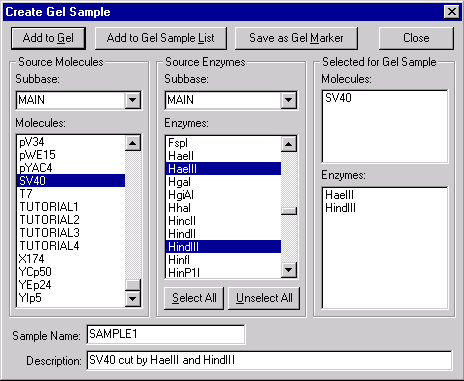
Press the Add to Gel button in the upper left corner of the dialog. Vector NTI cuts SV40 with the two RENs and adds the resulting fragments to the gel shown in the active Gel Display window. Press the Close button to close the dialog.
4. Add gel markers
Now let’s add two gel markers to provide yardsticks for the run. Click the Load Gel Marker button (  ) on the Window toolbar. A dialog box appears, showing the gel markers currently stored in the database.
) on the Window toolbar. A dialog box appears, showing the gel markers currently stored in the database.
Select the PBR322-HAEIII gel marker from the MAIN subbase and press the OK button. The gel marker is added to the second lane of the gel.
Click the Load Gel Marker button one more time. The Gel Markers dialog box appears again. This time select the SPP1-ECORI gel marker and press the OK button. The gel marker is added to the third lane of the gel.
5. Arrange the Gel Display Window conveniently
Now arrange the Gel Display window to your liking. It can be moved within the Vector NTI workspace using standard window manipulation techniques. You can move, resize, maximize, etc. Click the maximize box in the upper right corner of the Gel Display window. The window expands to fill the entire Vector NTI workspace.
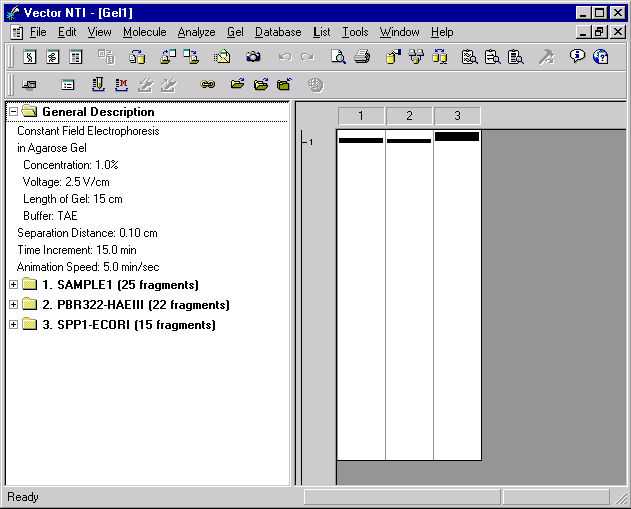
Note that the Gel Display window has a splitter dividing the text description of the gel (the text pane) from the graphical gel display (the graphics pane). You can drag the splitter to allow more space for either the text pane or the graphics pane. You can press the Switch Panes button (  ) in the Window toolbar to activate text pane or graphics pane.
) in the Window toolbar to activate text pane or graphics pane.
6. Inspect the Gel Display window text pane
With the Switch Panes button, activate the text pane of your Gel Display window. Now inspect the contents of the text pane. The pane holds several folders. The General Description folder describes the electrophoresis parameters. There is also a folder for each lane of the gel.
Double-click the folder labeled “1. SAMPLE1”. The folder opens, displaying a description of how the sample was obtained and subfolders for each fragment in decreasing order of length.
Double-click on the folder representing the longest fragment in the sample. This is the folder labeled 861. The folder opens, displaying information about the source of the fragment. For fragment 861, the source molecule is SV40, and the ends of the fragment are the HindIII site found at position 4003 and the HaeIII site found at position 4864. This information about the source of each sample is displayed in the Gel Display Window only. You can save samples in the database to be used later as gel markers. When you save the sample in the database, the information on the source of each sample is discarded, because for gel markers only fragment lengths are important.
Double-click the folder labeled “1. SAMPLE1”. This closes the folder.
Now double-click the folder labeled “3. SPP1-ECORI”. This is the gel marker found in lane 3 of the gel. The gel marker’s folder also has subfolders showing the size of each of its fragments. The longest fragment in SPP1-ECORI is 8000 bp. Double-click the folder labeled “8000”. The folder opens, revealing that it is empty. The folder is empty because for gel marker fragments, only the length is stored in the database.
7. Change display color for a fragment
You can use the text pane to change the color and style for displaying fragments. Double-click on the SAMPLE1 folder again, to open it. With the scroll bar, move down through the list of fragments until you see a folder labeled “540”. This represents a fragment with a length of 540 bp.
Place the mouse cursor over the “540” folder and press the right mouse button (Windows) or hold down the command key and click (Macintosh). A shortcut menu appears. Choose the Sample Fragment Properties command. A dialog box appears, allowing you to choose line style and color for this fragment. Leave the line unchanged, but change the color to green.
As the gel runs, you will now be able to locate the 540-bp fragment at a glance by looking for the green color.
8. Run the gel
We will now run the gel. Use the Switch Panes button (  ) to activate the gel pane.
) to activate the gel pane.
Press the Fit To Window button (  ), and the graphical gel display is sized to fit exactly within the current window.
), and the graphical gel display is sized to fit exactly within the current window.
Press the Step Forward (  ) button in the Window toolbar. The time indicator increases by fifteen minutes, and the fragments advance in the gel display. Press the Step Forward button several more times.
) button in the Window toolbar. The time indicator increases by fifteen minutes, and the fragments advance in the gel display. Press the Step Forward button several more times.
Press the Step Backward button (  ) in the Window toolbar. The time indicator and gel display back up.
) in the Window toolbar. The time indicator and gel display back up.
Now click once somewhere in the time indicator box to activate it. Enter “1:30” in the time indicator box, and press enter (Windows) or return (Macintosh). The gel display is set for that amount of elapsed time.
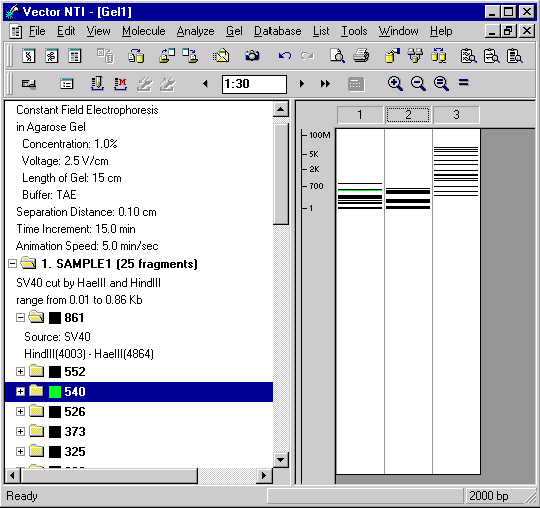
As the gel advances and retreats, watch how the scale keeps track of the fragment sizes.
9. Gel animation
Now let’s watch a continuous simulation of the gel run. Press the Animation button (  ) on the Window toolbar. The gel now runs continuously at the speed specified when you created the gel.
) on the Window toolbar. The gel now runs continuously at the speed specified when you created the gel.
When the gel time indicator reaches three or four hours, press the Animation button again to stop the run. The fragments are now well spread out.
10. Inspect the Gel Display window’s graphics pane
Now let’s inspect the Gel Display window’s graphics pane in more detail. You can zoom the graphics pane in and out to show more or less detail. Press the Zoom In button (  ); note how the picture was magnified. Press the Zoom In button again several times until the narrow lines representing the fragments change to fuzzy gray bars. Use the scroll bar at the right side of the graphics pane to move up and down the gel when the magnification is high.
); note how the picture was magnified. Press the Zoom In button again several times until the narrow lines representing the fragments change to fuzzy gray bars. Use the scroll bar at the right side of the graphics pane to move up and down the gel when the magnification is high.
Press the Zoom Out button (  ) several times until the gel is fully visible in the graphics plane.
) several times until the gel is fully visible in the graphics plane.
Press the True Scale button (  ). The graphical gel display is sized to approximately its true scale in the real world.
). The graphical gel display is sized to approximately its true scale in the real world.
Now press the Fit To Window button (  ) again, and the graphical gel display is sized to fit exactly within the current window.
) again, and the graphical gel display is sized to fit exactly within the current window.
11. Zoom in on the gel’s graphical display
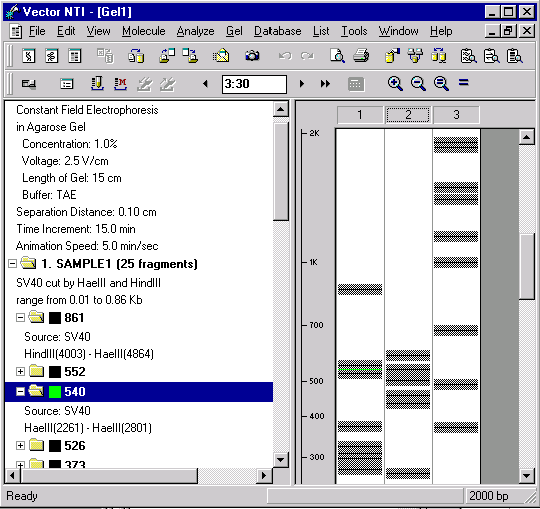
Now let’s look at the gel with high magnification. Press the Zoom In button several times, until gray bars appear around the lines representing the fragments. Using the vertical scroll bar, move down along the gel until your green fragment appears.
The gray bars show the minimum distance by which fragments must be separated in order for you to distinguish them in the gel.
Note that the gray bars of many fragments touch each other. This indicates that you will not be able to distinguish those fragments in the gel. The size of the gray bars is determined by the Separation Distance parameter, which is set at the time the gel is created. You can change the Separation Distance parameter with the Gel And Display Setup button (  ) of the Window toolbar. This allows you to set your own distance conditions for fragment separation.
) of the Window toolbar. This allows you to set your own distance conditions for fragment separation.
12. Change to a polyacrylamide gel from the Electrophoresis Profile
Vector NTI lets you store electrophoresis parameters in a Electrophoresis Profile. This way you can define values that are most convenient for you and recall them easily.
Let’s change the parameters of the gel we are running by selecting a new data set from Vector NTI’s Electrophoresis Profile.
Click on the Gel And Display Setup button (  ) in the Window toolbar. A dialog box appears, allowing you to change the electrophoresis parameters.
) in the Window toolbar. A dialog box appears, allowing you to change the electrophoresis parameters.
Click the down arrow on the Electrophoresis Profile box, and a list of available sets of parameters appears. Select the Example of Polyacrylamide Gel, and press the OK button. The electrophoresis parameters in the active Gel window are changed.
When you change the electrophoresis parameters, the contents of the lanes of the gel are not affected.
Now zoom in and scroll through the gel again and note the changed positions of the various fragments.
13. Estimate fragment separation time
Some fragments in our gel are still too close together to distinguish. (These are fragments whose gray zones overlap.) Let’s use Vector NTI’s built-in calculator to estimate how long the gel would have to run to separate some of these fragments.
Use the Switch Panes button to activate the graphics pane. Click the mouse in the time indicator box of the Window toolbar, enter two hours and thirty minutes (2:30), and press enter (Windows) or return (Macintosh).
Press the Fit to Window (  ) button to set the gel to its standard size.
) button to set the gel to its standard size.
Press the Zoom In button (  ) three times to magnify the view of the gel.
) three times to magnify the view of the gel.
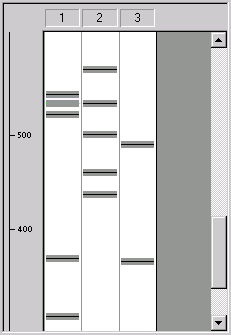
Look at the first lane of the gel, containing SAMPLE1. With the scroll bar, scroll down until you find the section of the gel containing fragments with lengths between 500 and 700 bp. You will see three fragments just above the 500-bp mark. The middle fragment (length 540 bp) will be green, because you set its display color in a previous step of this tutorial. The fragments’ gray borders overlap with each other, indicating that these fragments are too close together to be distinguishable on the real gel.
Move the mouse cursor until it is within lane 1 of the gel display. The cursor will change to a horizontal I-beam (  ). Place the mouse cursor above the three fragments. Press and hold the left mouse button, and move the mouse downwards. A highlighted band appears as you move the mouse. This is the selection zone. Select the three unseparated fragments by highlighting them with the selection zone.
). Place the mouse cursor above the three fragments. Press and hold the left mouse button, and move the mouse downwards. A highlighted band appears as you move the mouse. This is the selection zone. Select the three unseparated fragments by highlighting them with the selection zone.
Now press the Calculator button (  ) in the Window toolbar. A message appears, telling you that the gel would have to run for 5 hours and 30 minutes to separate the fragments you selected. The time calculation is an estimate and is intended to give you a general idea of what to expect from your real-world gel runs.
) in the Window toolbar. A message appears, telling you that the gel would have to run for 5 hours and 30 minutes to separate the fragments you selected. The time calculation is an estimate and is intended to give you a general idea of what to expect from your real-world gel runs.
Using the vertical scroll bar, move down along the gel until the selection appears. Click anywhere in the gel pane to remove the selection.
14. Save the Gel Display window to a Gel Document file
To save all the information contained in the Gel Display window, including gel settings and fragments positions, you should save the contents of the Gel Display window in a Gel Document file. Let’s save the current contents of the Gel window. From the Gel menu, choose the Save as Document command.
A dialog box appears, asking you for a filename. Name the file “Tutorial.gd” (Windows) or “Tutorial Gel” (Macintosh) and press the OK button (Windows) or the Save button (Macintosh). The gel is saved in a document file.
Let’s close the Gel window. From the Gel menu, choose the Close command.
Now open the Gel Document you created. From the Gel menu, choose the Open Document command, and select the name of the saved document file from the file list. Your Gel Document opens, and all your fragments and settings have been preserved.
14. Close the Gel Display window and exit Vector NTI
You have now taken a first look at Vector NTI’s Gel Display windows. Close the window using the Close command in the Gel menu, and then quit the program with the File menu’s Exit command (Windows) or Quit command (Macintosh).
This concludes the Gel tutorial.
See Also:
Chapter 12 -- Gel Display Windows
Introduction
Chapter 11 -- Display Windows and User Interface
Chapter 6 -- PCR Analysis
Chapter 7 -- Molecule Construction
Chapter 1 -- Display Windows
Chapter 18 -- Reports Generation
Back to Index