- Overview
- Installation
- Use of this manual
This Chapter describes an overview of the functions that you can perform with the Vector NTI Software. Details on how to use the program can be found in the Vector NTI Tutorials or User’s Guide. You can use the Index at the end of the Manual to find specific topics quickly. However, we strongly recommend that you read this chapter first to get an overall appreciation of the program.
The Program Structure
Vector NTI is a software package to aid molecular biologists, genetic engineers, students, and others in the study, manipulation and creation of biological molecules.
The logical structure of Vector NTI is shown below:
Database
Vector NTI database contains DNA/RNA molecules, protein molecules, enzymes (restriction endonucleases), oligonucleotides (including PCR primers, sequencing primers and hybridization probes), and gel markers. Vector NTI has a special functional module called “Database Explorer” to deal with all the types of database objects. The DB Explorer allows you to create new objects, edit and delete old objects, perform database searches for interesting objects, organize objects into convenient groups (subbases), import/export objects, and create Vector NTI “archives” of objects to share with other users.
Vector NTI also has two additional data managers: 1) A Contact Manager that allows you to create, modify, and organize the address data of persons or organizations you are frequently communicating with, and; 2) A User Field Manager that allows you to enter your own data fields which you can define and use to describe your database objects.
There are three general functions dealing with the entire database: Database Backup, Database Restore, and Database Cleanup. The Database Backup function makes a copy of your database in the specified subfolder of your hard or floppy disk. The Database Restore function restores all database files from the desired backup. The Database Cleanup function allows you to compress the database by removing all unnecessary data from the datafiles and database tables. These data may accumulate as a result of extensive creating and deleting of database objects. Although these data do not affect how Vector NTI functions, we recommend that you cleanup your database once a week. The Database Cleanup can also be used as a recovery function when database files and/or tables are lost or damaged. All three general database functions are available from the Vector NTI Database menu.
Creating new molecules
There are four different ways of creating new DNA/RNA and protein molecules:
- Import. You can import molecules (including their feature tables) from GenBank/GenPept, EMBL/SWISS-PROT and FASTA formats. You can also import nucleotide or amino acid sequence from an ASCII file of flexible format, and Vector NTI will automatically create the new database molecule and assign the sequence to the molecule. Let us note that Vector NTI allows you to import molecule data even from “quasi-GenBank” files (e.g., GCG’s “GenBank” output files).
- Creating new molecules “from scratch.” You can define a molecule’s nucleotide or amino acid sequence by hand or paste it from the clipboard using Vector NTI sequence editor and save the new molecule to database.
- Construction. New DNA/RNA molecules can be created from user descriptions of compatible component fragments (e.g., fragments of other molecules). This process is called construction in Vector NTI terms.
DNA/RNA molecules can be composed of several different types of fragments: “dummy” fragments (pieces of DNA of unknown nucleotide sequence), short (up to 120 bp) nucleotide sequences, linkers, adaptors, and fragments of molecules from Vector NTI database. After being constructed, a molecule is integrated into the database and may participate in all further operations and analysis. For instance, it may be used for the construction of new molecules.
Vector NTI allows you to define the fragments’ termini in different ways and to modify the termini using all known biochemical operations. Unlike other programs, Vector NTI checks the compatibility of the fragments in all possible situations (e.g. non-palindromic sites).
- Translation. New protein molecules can be created by translation of a coding region of an existing DNA or RNA molecule.
- Design. The new DNA/RNA molecules can be created with the aid of Vector NTI’s built–in biological knowledge. This process is called design. In design, the user compiles a list of fragments which will describe a goal molecule. Instead of personally choosing restriction sites, methods of terminus modification, etc., the user leaves all choices up to Vector NTI. Using built-in biological knowledge, Vector NTI creates a construction plan that takes advantage of the best possible restriction sites and genetic engineering techniques.
Component fragments describing the goal molecule for design have a more subtle nature than the fragments used for construction. For design, the goal molecule must consist of exactly one recipient fragment and one or more donor fragments. You may allow Vector NTI to choose all the restriction sites itself or you may indicate a specific restriction site at one or both ends of recipient or donor fragments. Moreover, for the recipient fragment you may request Vector NTI to save or lose such a specific restriction site. You may also request to delete a region of specified length from the recipient molecule during the cloning process.
The purpose of donor fragments is to carry functional signals into the recipient. When you describe a donor fragment you usually do that not by the fragment’s termini but by the functional signals it contains. You also may indicate the maximum flanking regions which you are willing to permit for these functional signals.
One important feature of the molecule design process is that you have many options to “adjust” Vector NTI actions according to your specific needs. You may permit or prohibit specific genetic engineering techniques such as use of partial digests, use of linkers, adaptors, etc.
Sequence and feature map of new molecule
When a molecule is imported from GenBank/GenPept, EMBL/SWISS-PROT or FASTA file, its sequence and feature map (if any) are converted from the file. You may edit the sequence and the feature map visually in a Molecule Display window for the molecule or using the appropriate built-in Editors. It may be particularly useful for assigning or changing the name of a molecule’s feature. Of course, you may add new features to a molecule’s feature map.
When a molecule is created “from scratch” it has no feature map and sequence. You have to enter those data yourself.
When a DNA/RNA molecule is created during the “design” or “construction” process, its nucleotide sequence and feature map are created automatically from its parents. You may add new functional signals to the molecule’s functional map. You may also delete or edit automatically generated functional signals or nucleotide sequence. If you delete or edit the signals or sequence, the molecule will become disconnected from its parents to avoid data inconsistency. In this case, the molecule will keep all its data, excluding the component fragments. The system will prompt you if such a situation occurs.
When a protein molecule is created by translation of a coding region of a parent DNA molecule, its sequence is generated automatically. If you edit the sequence, the protein molecule will become disconnected from its parent to avoid data inconsistency.
Restriction maps of DNA/RNA molecules
Vector NTI doesn’t store restriction maps of sequenced parts of DNA/RNA molecules in the database. These maps are created “on the fly” whenever you need them. But for the unsequenced molecule regions, you may enter the known positions of restriction sites. Then the molecule’s nucleotide sequence will be automatically modified so that these sites will participate in the creation of a restriction map. Moreover, that information will be inherited by all the molecule descendants. Vector NTI automatically checks the compatibility of the manually entered restriction sites (e.g., when they overlap), and their consistency with the molecule’s nucleotide sequence.
Creating new enzymes, oligos, and gel markers
Enzyme objects may be created:
- “From scratch,” using Enzyme Editor.
- By importing them from REBASE database.
Oligonucleotide objects may be created:
- “From scratch,” using Oligo Editor.
- From the results of searching for PCR primers, sequencing primers and hybridization probes.
Gel marker objects may be created:
- “From scratch,” using Gel Marker Editor.
- By cutting the database molecules by restriction endonucleases.
Editing database objects
The Database Explorer allows editing of existing database objects using the appropriate Editor dialog. Probably, for the database molecules you will find it more convenient to edit information in the Molecule Display windows (see below), which allow you to immediately visualize editing results.
Database search
Vector NTI allows searching of the database using the following types of data as search conditions:
For DNA/RNA molecules:
- Form (circular/linear)
- Storage type (basic or constructed from other molecules)
- DNA/RNA
- Replicon type
- Extra–chromosome replication capabilities
- Size
- Description/comment/user fields text
- Keywords
- Ancestors
- Functional signals (including such conditions as “any promoter,” “CDS AP(R) or TC(R),” etc.)
- Oligonucleotides and short peptides with a specified homology to a molecule
For protein molecules:
- Storage type (basic or translated from DNA molecule)
- Size
- Description/comment/user fields context
- Keywords
- Ancestors
- Features (including such conditions as “any transmembrane region” etc.)
- Short peptides with a specified homology to a protein
For enzymes:
- Length of recognition site
- Description context
- Keywords
- Nucleotide substring in recognition site
- Ambiguity
- Palindromes/Non-palindromes
For oligonucleotides:
- Description context
- Keywords
- DNA/RNA
- Nucleotide substring in oligonucleotide
For gel markers:
- Description context
- Keywords
- Fragment length limits
Parent-Descendant connections
Vector NTI is able to manage parent-descendant connections between molecules. Imagine that the DNA molecule NEWMOL1 is constructed from two fragments: the fragment of molecule A and the fragment of molecule B. Then imagine that the molecule NEWMOL2 is also constructed from two fragments: the fragment of NEWMOL1 and the fragment of molecule C. Now we have the following parent-descendant connections between A, B, C, NEWMOL1, and NEWMOL2:
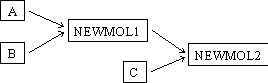
- A and B are the “direct ancestors” of NEWMOL1
- NEWMOL1 and C are the “direct ancestors” of NEWMOL2
- A and B are the “ancestors” of NEWMOL1
- A, B, NEWMOL1, and C are the “ancestors” of NEWMOL2
- NEWMOL1 is the “direct descendant” of A and B
- NEWMOL2 is the “direct descendant” of NEWMOL1 and C
- NEWMOL1 is the “descendant” of A and B
- NEWMOL2 is the “descendant” of A, B, NEWMOL1, and C
Vector NTI also treats a protein molecule constructed by translating a coding region of a DNA molecule as the “direct descendant” of the DNA molecule.
Parent-descendant connections are important data of the molecule database. They allow you to study the “tracks” of constructs in your database and to pass the changes automatically from parents to descendants. However, the presence of such data may cause problems in molecule management. These are discussed below.
Imagine that you change the nucleotide sequence of DNA molecule A so that the change occurs in the fragment used for constructing NEWMOL1. If the sequences of NEWMOL1 and NEWMOL2 remain unchanged, the data of the database will become inconsistent. Since molecule NEWMOL1 contains a fragment of A, the sequence of this fragment in NEWMOL1 must be the same as the new sequence of A. However, if the sequence of this fragment of NEWMOL1 is the old sequence of A, then this is different from the actual A sequence and therefore NEWMOL1 and A are not consistent. Alternatively, imagine that you deleted A from the database. The next attempt to reconstruct NEWMOL1 will fail because Vector NTI will not find one of the parent molecules. There are a lot of other situations where one should be careful when dealing with molecules which have parent-descendant relationships.
Fortunately, Vector NTI automatically takes care of such situations. When you are trying to change an existing database molecule, Vector NTI automatically checks whether the molecule has descendants. If the molecule has descendants and the changes may affect them, Vector NTI gives you the choice to pass the changes through all the descendants or to disconnect the direct descendants. Also, when you are deleting a molecule which has descendants, Vector NTI automatically disconnects the direct descendants. So, you can enjoy the presence of parent-descendant information and at the same time not worry about possible problems, because Vector NTI does all necessary work to avoid data inconsistency.
Archives—a tool for sharing database information
Vector NTI allows you to place your molecules, enzymes, oligonucleotides and gel markers into archives—documents of special format. These archives can be transferred to another computer (Mac or PC) and read by Vector NTI. So, archives give you the possibility to share you objects (for instance, your own molecule constructs) with your colleagues, or to have them simultaneously on several computers (for instance, at work and at home). There are two important points regarding Vector NTI archives. The first is that all the molecule information will be archived including component fragments (if the molecule is constructed from other molecules) and parent-descendant connections between the molecules. The second is that Vector NTI automatically checks the consistency of a molecule archive. Imagine that you place a number of your constructs into an archive to give them to your colleague but forget to include some of the parents of these constructs. Your colleague may have problems dealing with them. If she, for instance, wants to reconstruct some of your molecules, the program will not find the necessary parents in his database. But Vector NTI will check this (among other possible situations ) at the stage of archive creation and allow you to solve the problem by adding necessary parents or disconnecting them. Also when the archive is loaded into a new database, Vector NTI will check if it is affecting any database molecules and recalculates them if necessary.
Subbases—convenient groups of database objects
You may organize your DNA/RNA molecules, protein molecules, enzymes, oligos, and gel markers into an arbitrary number of subbases. Each subbase is a named group of corresponding objects. For instance, you may have one subbase for each of your molecule families, one for each taxonomic group, etc. Subbases are very useful when you need to perform an operation on several objects at once—for instance, to delete them, or to place them into a Vector NTI archive and transfer them to another computer. Subbases are also a powerful tool for restricting the number of objects under consideration—when you perform a database search or select an object from a set of database objects. Vector NTI Database Explorer has a number of convenient commands for creating and managing subbases of all database objects.
Organizing your biological objects
Vector NTI provides the user with a unique possibility to organize conveniently and to manage effectively a large scope of molecular biology data. All Vector NTI database objects—DNA/RNA molecules, protein molecules, enzymes, oligonucleotides and gel markers—have description fields allowing you to specify any object property in formal and semiformal ways. These fields are “Description” which is a single line of arbitrary text, “Comment” which is a text of arbitrary length, “Keywords” which is a list of short text strings, and possibly a number of user-defined fields which may have different format. Vector NTI allows you to search through a database using subbases, keywords and text search for database objects of any type. So, even if you have thousands of objects (e.g., constructed molecules) in your database, you cannot lose them. Grouping the objects by subbases, assigning keywords to them, and using recognizable text fragments in their descriptions, comments or user-defined fields, you can get the objects of particular interest in a second and work directly with them. So, in addition to Vector NTI’s other functions, you can treat the program as a global “organizer” for your biological data.
Molecule Display windows
Molecule Display windows are Vector NTI’s mechanism for inspecting and editing text description, graphical maps and sequences of DNA/RNA and protein molecules. They are extremely powerful and convenient graphical tools, allowing you to study and edit molecules stored in Vector NTI database, create Vector NTI Molecule Documents, prepare publication-quality figures and visually select fragments of DNA/RNA molecules for molecule construction and design. A Molecule Display window can show an entire molecule or a selected region of molecule. This is useful if you want to manipulate a small region of a larger molecule.
Viewing the molecules
Molecule Display windows consist of text, graphics and sequence panes. A Display window’s text pane contains a general molecule description and the following data:
For both DNA/RNA and protein molecules:
- A Feature map in text form. You may select the types of features to be shown choosing them from the list of approximately 60 GenBank-compatible types for DNA/RNA molecules and more than 100 for protein molecules.
For DNA/RNA molecules:
- A Restriction map in text form. You may select any number of restriction endonucleases. Also you may filter the results of restriction sites search specifying permitted terminus types, maximum number of recognition sites, etc.
- ORFs found on the molecule. You may set the minimum ORF size, define your own start and stop codons, choose the possibility of finding nested ORFs (ORFs that have the same stop codon but different start codons), and switch between different coordinate systems for defining ORF phases.
- Results of a motif search. You may search for motifs on direct or complementary strand of DNA or on both strands with a specified homology between the motifs and molecule sequence. The desired homology can be described as a minimum homology threshold or just as the requirement to find “the best” homology for a particular motif. You may choose oligonucleotides from the Vector NTI database and use them as motifs. Also you may filter the results of motifs search, specifying the permitted maximum number of recognition sites and other parameters. In case of an ambiguous motif, the system allows you to choose between minimum, maximum or average homology as a criteria for the motif’s “closeness” to the molecule’s sequence. In the results of an ambiguous motif search all three homologies are shown. In addition, you can require 100% homology for a motif’s 3’ end of specified length. This may be useful, for instance, when you are trying to check the applicability of your own PCR primers.
- PCR primers, sequencing primers and hybridization probes found in the primer analysis. A Molecule Display Window allows you to not only study the primers and their physico-chemical characteristics, but also to manipulate them in a convenient manner: you may immediately analyze the primers with the comprehensive Vector NTI function of oligo analysis, save the primers into the oligo database, etc. Also, you may easily view a PCR product by creating its separate Display window with one click of mouse button.
- A “Construction plan” which is the result of Vector NTI molecule design process (if the molecule was designed using Vector NTI built-in biological knowledge). The plan contains descriptions of all the necessary cloning steps. For each cloning step, Vector NTI chooses the optimal restriction sites and methods of termini modification (if necessary), describes the ligation types, recommends preselection methods and transformation system, describes in detail an appropriate method for clone analysis after transformation (e.g. provides the most convenient restriction sites for restriction analysis and primers for PCR analysis) and lists important restriction sites for the resulting molecule.
For protein molecules:
- Analysis results in tabular form. The results include molecular weight, molar extinction coefficient, isoelectric point, etc.
- A Molecule Display window’s graphics pane contains a circular (for circular DNA/RNA molecules) or linear molecule map. Vector NTI uses the original advanced layout algorithms to place the graphics objects optimally. All graphics objects are movable, resizable and editable.
A Molecule Display Window’s sequence pane contains the formatted nucleotide or amino acid sequence of a molecule. For DNA/RNA molecules, Vector NTI can show restriction sites, motifs, ORFs and functional signals together with the sequence and translate the sequence in any reading frame. For protein molecules, Vector NTI can display features together with the amino acid sequence.
You can print any Molecule Display Window’s pane (printing output quality is limited only by the resolution of your printer), or you can save it to the external file (Windows Metafile format on Windows and PICT format on Macintosh) or put it to the Clipboard. The Clipboard allows you to insert the maps, sequences and molecule descriptions directly into your documents prepared in your favorite word processor.
You can link text and graphics panes of a Molecule Display window to manage them more effectively. This is extremely useful in the case of maps of large molecules containing hundreds of features and restriction sites.
Editing the molecules
Molecule Display windows allow you to edit your molecules and immediately visualize the results of editing. You can modify a molecule’s sequence by inserting, deleting and replacing sequence fragments. You can edit a molecule’s general data such as description, comments and keywords, edit, delete and add features, and insert restriction sites into the non-sequenced DNA/RNA molecule regions. Also you can perform more general operations on the molecules directly from the Display windows: change a molecule’s starting coordinate, export a molecule into an external file, etc. Changes you make on the molecules are immediately reflected in the Molecule Display windows so that you can view them and decide whether or not you would like to keep them. When your editing is complete you can save the molecule into the database to make the changes permanent.
Using Molecule Display windows for construction and design of new molecules
A Molecule Display window’s graphics pane has an operational mode that allows you to define DNA/RNA molecule fragments for insertion into new molecules directly on a graphics map. You can easily “cut” DNA molecules by restriction enzymes, place the fragments into the Molecule Goal List, and construct the new molecule from defined fragments by pressing one button. You can even describe more complicated fragments used in Vector NTI’s molecule design process: functional signals with limited flanking regions, fragments with approximate boundaries and so on. Vector NTI uses a convenient Fragment Wizard dialog to help you easily describe the molecule fragments of different types in an appropriate way.
You can also easily construct a new protein molecule by translating the selected fragment of a DNA or RNA molecule. The translated protein molecule will be automatically saved into the database and opened in a separate Molecule Display window.
Creating Molecule Documents
You can save all the information from a Molecule Display window to a permanent storage by creating a Molecule Document file. A Molecule Document is an extended GenBank (for DNA/RNA molecules) or GenPept (for protein molecules) file containing all the molecule data for which there are appropriate GenBank/GenPept fields, such as feature table or nucleotide sequence. Additional information in the COMMENT fields describes not just the molecule but the entire Molecule Display window which was saved into the document. Since the Molecule Document format is based on GenBank/GenPept formats, not only Vector NTI, but any program able to import GenBank/GenPept files can also import Vector NTI Molecule Documents (all the Vector NTI-specific information will be ignored ). Another copy of Vector NTI or public domain Vector NTI Viewer can open a Molecule Document into a Molecule Display window which will contain all the information the original window contained and look exactly the same as the original window. So, you can share with your colleagues large information fragments encapsulating not just the data of a molecule but a wide range of results of your work with the molecule, such as component fragment or a design plan description of your construct, result of ORF and motif searches, PCR analysis and so on. Moreover, by manipulating Display window panes, editing graphics maps and inserting annotations you can emphasize the most important aspects of your work reflected in the Molecule Display window. With the concept of Molecule Documents, Vector NTI offers a new type of communications between biologists, allowing them to exchange the results of their work in the specialized electronic form.
Internet Tools
Vector NTI belongs to a new generation of Internet-friendly applications that allow you to share your data and results of your work with your colleagues around the world and to use remote bio-services for analyzing your data. Every time you create a Molecule Document, Vector NTI offers to create the accompanying HTML file containing local HTML link to the document, so you can upload both files to your WWW site and make your document visible to the whole world. You can also export a group of your molecules, e.g. your own constructs, or a group of your oligonucleotides, e.g. your own PCR primers, and Vector NTI automatically creates an HTML file containing your molecule or oligo names as HTML links. Using these possibilities you can quickly prepare data for publishing on a WWW site and make them immediately available to everyone not having any special WWW-related knowledge. Moreover, now you can not only publish your data but also analyze them using a vast amount of analysis resources available worldwide. You can send your data to several Internet servers providing different kinds of analysis, database search and alignment directly from Vector NTI. You can also add new connections to Internet servers and local programs yourself by using Vector NTI’s “scripts” mechanism. When new scripts become available, you will be able to download them from the Vector NTI WWW home page. With this concept of connectivity, Vector NTI brings new standards into the area of scientific information exchange.
Oligo and Primers
Vector NTI can find PCR primers, sequencing primers and hybridization probes for any desired part of a DNA molecule. The primers and hybridization probes can be saved to the Vector NTI’s oligo database. You may then use them as your own primers and probes in further primer analysis, e.g. checking the usability of your own primer or finding the best primer pair (for PCR) for different molecules and under different conditions.
You may set up many parameters and restrictions for desired primers during primer analysis. Among them: melting temperature, length, % GC, 3’ end parameters, palindromes, nucleotide repeats, dimers and hairpin loops, etc. You may adjust the relative “importance” of primer parameters, creating your own scoring functions for determining “the best” primers. In the case of PCR primers you may attach short nucleotide sequences (e.g. restriction sites) to the primers’ ends and analyze those combined primers. You may also require all the primers and hybridization probes to be unique on the molecule with a specified homology threshold to avoid cross-hybridization.
From many places within Vector NTI you may call up the Oligo Analysis dialog box to analyze oligonucleotides (including primers found in an analysis). You can also generate and check all possible duplexes between any two oligonucleotides, e.g. between sense and antisense PCR primers. The results of oligo and duplex analysis may be saved in an external text file.
Let us emphasize that Vector NTI, unlike other software, is very accurate when dealing with degenerate primers. It concerns calculating both primer homology and thermodynamic parameters. Using Vector NTI for primer search and analysis you can be sure that you find the optimal solution even in the most difficult cases.
Gel Display windows
Gel Display windows are Vector NTI’s powerful graphical tools for supporting your electrophoresis work. Gel Display windows may be created for different types of electrophoresis including pulse field electrophoresis. You may add one or more Gel Samples and Gel Markers (see below). You can then run your gel. In the Gel Display window’s text pane you can view general electrophoresis data or specific fragments. You can also assign colors and line formats to individual fragments to distinguish them from other fragments. The Gel Display window’s graphics pane shows you a model of what the gel would look like in real life. You can zoom the display in or out, fit it to the size of the Gel Display window, or view the gel in approximate true scale. You can also “mix” Gel Samples through the special data clipboard called Gel Sample List.
Creating Gel Display windows
To create a new Gel Display window you have to define its parameters. You may do that by choosing your favorite parameters from the Electrophoresis Profile, or by describing a new one by setting the following data:
- electrophoresis type: agarose gel with constant electric field, polyacrylamide gel with constant electric field, or agarose gel with pulsed electric field
- buffer type
- electrophoresis parameters: gel concentration, applied electric field, gel length, and pulse time for the pulse field electrophoresis
- display parameters
You can change the settings of a Gel Display window at any time after creation. This allows you to “adjust” your gel for better separation of the molecules’ fragments by varying gel concentration, applied voltage, etc., or even trying to use a different mode of electrophoresis.
Creating Gel Samples and Gel Markers
In Vector NTI, gels contain two types of data: Gel Samples and Gel Markers. Gel Samples are sets of particular fragments of particular molecules. They may be created by cutting database molecules by restriction enzymes with the aid of Vector NTI’s convenient interface. You may also add any particular molecule’s fragment to a Gel Sample directly from the Molecule Display window. Gel Samples are not stored in the Vector NTI database but are created on the spot for short-term use. If a Gel Sample is saved to the database, it becomes a Gel Marker.
Gel Markers are sets of fragments of known lengths that are not connected to any particular molecule from the Vector NTI database. They are used as yardsticks for measuring migration speed. Gel Markers are stored and loaded from the Vector NTI database. They may be either created from Gel Samples, or directly using Vector NTI’s Gel Marker Editor.
Running the Gel
When a gel Display window is created, and your samples and markers added to the window, you can run a gel that models your real life experiments. You can choose your time increments and step forward or backward through the run in those increments, or choose the automatic run mode and see the animation of the gel. You also can set the specific length of time for your gel run and see what happens with your fragments in the gel after that time. You can define “separation distances” for your fragments to better visualize whether your particular fragments have been separated, and you can link text and graphics panes of the Gel Display window to show or hide individual fragments for more convenient studying of the gel picture. Finally, you may ask Vector NTI itself to calculate what time is required for separation of any number of fragments selected in your gel.
Installation
Installing Vector NTI for Windows
Before installation of Vector NTI for Windows, make sure that Windows (Windows 95 / Windows 98 / Windows NT version 4.0 or later ), is properly installed on your computer. To install Vector NTI for Windows, insert installation disk #1 in a floppy drive (or CD ROM into the CD drive), go to the Start menu of Windows 95, Windows 98, or Windows NT, and select Run. A dialog box will appear asking you what program to run. Enter the letter of the drive the first installation disk is in, followed by a colon and the word setup, and then the enter key. For example, if you were installing from drive a:, you would type a:setup and press enter. Vector NTI will install itself on your hard drive, and you can then launch the program by double-clicking its icon in the Start menu or the appropriate Windows Explorer folder.
Installing Vector NTI for Macintosh
To install Vector NTI for Macintosh, you must have Apple System software, version 7.0 or later, already installed on your Macintosh. Insert the disk #1 into the floppy disk drive (or CD ROM into the CD drive). Start installation process by double-clicking Install Vector NTI icon. When installation program starts, follow the instructions on the screen. You can start Vector NTI from within the Finder by double-clicking its icon in the folder in which you installed Vector NTI.
Initializing Vector NTI Database
When you start Vector NTI for the first time after installation, Vector NTI creates an empty database and asks you a permission to import initial set of database objects from Vector NTI archives installed in Vector NTI working folder or directory. Press the OK button in a confirmation dialog. The DNA/RNA molecules, proteins, enzymes, oligos, and gel markers will be imported to form the initial Vector NTI database.
Use of this manual
The Vector NTI manual is divided into two parts—Tutorials and User’s Guide. The Tutorials, in chapters 1–10, give you a hands-on introduction to the program. The User’s Guide in chapters 11–17 describes the program’s functions in detail.
We hope you enjoy InforMax’s Vector NTI!
See Also:
Chapter 13 -- Oligonucleotides and Primers
Chapter 2 -- Molecule Operations
Chapter 4 -- Tools and Internet Connectivity
Chapter 5 -- Database Explorer
Chapter 11 -- Display Windows and User Interface
Chapter 15 -- Database Operations
Chapter 17 -- Vector NTI Tools
Chapter 7 -- Molecule Construction
Chapter 8 -- Molecule Design
Chapter 14 -- Molecule Components and Construction
Chapter 1 -- Display Windows
Chapter 9 -- Advanced Molecule Design
Chapter 6 -- PCR Analysis
Chapter 18 -- Reports Generation
Chapter 16 -- Molecule Design
Chapter 3 -- Working With Graphics Representation
Chapter 10 -- Working With Gel Display windows
Chapter 12 -- Gel Display Windows
Back to Index