- Background
- Your Task: Using Vector NTI to send data to WWW servers
This chapter introduces you to Vector NTI’s Tools, a powerful mechanism for extending Vector NTI’s functionality and providing connection to external programs and Internet services.
Your Task: Using Vector NTI to send data to WWW servers
Your task will be to send data from Vector NTI to several public WWW servers, practicing simple BLAST search, alignment and analysis. Follow the steps in the order shown. Figures show what your screen should look like at various points along the way.
1. Launch Vector NTI
Launch Vector NTI by double-clicking its icon in the program group or folder in which you installed Vector NTI and activate the workspace window by clicking on its title (Windows) or status bar (Macintosh).
2. Open pBR322 in a new Display window
Click the Open Nucleic Acid from Database button (  ) on the main toolbar to open a new Molecule Display window.
) on the main toolbar to open a new Molecule Display window.
A dialog box appears, asking you to select a molecule to be opened in a new Display window. Select the MAIN subbase and the pBR322 molecule, then press the OK button. A Molecule Display window opens, containing pBR322’s text description, sequence, functional and restriction maps.
3. Select the whole sequence of pBR322 and use the BLAST Search tool
Click anywhere inside the the pBR322’s Display window to activate it. Now, choose the Compare Against4
GenBank via BLAST on NCBI Server command from the Tools menu.
Note Some World Wide Web browsers require the Internet connection to be established before you start the browser. If your TCP/IP stack or other Internet connection software can not be started “on the fly” you need to connect before selecting any of Vector NTI’s Internet-related commands.
Vector NTI shows the Sequence Data dialog allowing you to choose the range and strand of the sequence to be sent to the server:
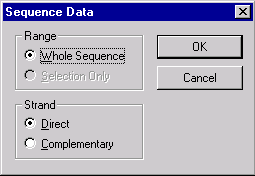
Choose Whole Sequence / Direct Strand and press OK to continue.
When you execute any Internet-related command for the first time, Vector NTI tries to configure its connection to the WWW browser before proceeding. Select the name of your WWW browser from the list and press the OK button to continue.
Note The standard Vector NTI distribution contains support modules for many popular WWW browsers. If your browser is not in the list, try selecting one of the “Autodetect” modules from the displayed list. The latest additions to the list are available on the Vector NTI WWW home page:
http://www.informaxinc.com/
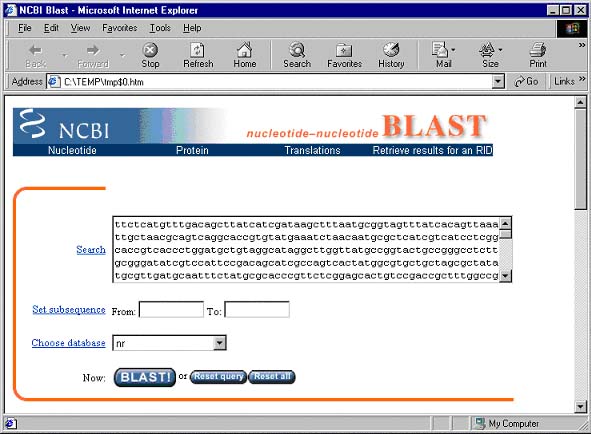
This page is Vector NTI's gateway to the NCBI BLAST Search server and contains options that can be used to change various BLAST search parameters.
The sequence of pBR322 is already entered in the Search field. In the Choose database section, select the nr database. Scroll to the bottom of the page. If your email address appears in the Send results by e-mail text box, delete it. Press the BLAST! button.
Note At this step, your WWW browser may display a dialog box or start a program to establish an Internet connection. If this happens, enter the required parameters and continue.
Once an Internet connection with the NCBI server is established, you should see the NCBI formatting BLAST page. To proceed, press the Format! button on that page. The response time for the BLAST results varies depending on the server load. If the server is busy, wait several minutes and resubmit the query.
Note Since the interface of WWW servers changes from time to time, you may receive a message telling that the requested resource is not found. This means that Vector NTI’s server gateway page is obsolete. The latest versions of Vector NTI gateway pages are available on the Vector NTI WWW home page.
When you receive the results from the BLAST server, click on the second molecule link (at the time of this writing, the first hit was pBR322 itself and second hit was U03501/YRP7 ). Another page appears, containing the GenBank description of the molecule:
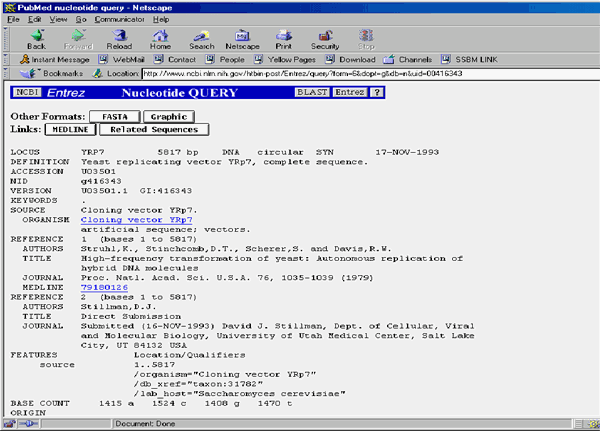
4. Display the result of the query in a Document window
Now let’s transfer the result of the query back to Vector NTI. Choose the Select All command in the Edit menu of your browser (or, if your browser does not have the Select All command, select the contents of the window with the mouse) and use the Copy command from the Edit menu (or its analog in your browser) to copy the selected text to the clipboard. Your selection should include all GenBank text; Vector NTI will ignore all extra text before the start of GenBank data (LOCUS line) and after the end of the data (//). Switch back to the Vector NTI workspace and select the Open4
GenBank From Clipboard command from the Tools menu. Vector NTI opens your molecule in a new Display window, automatically generating the restriction map and graphics representation of the molecule.
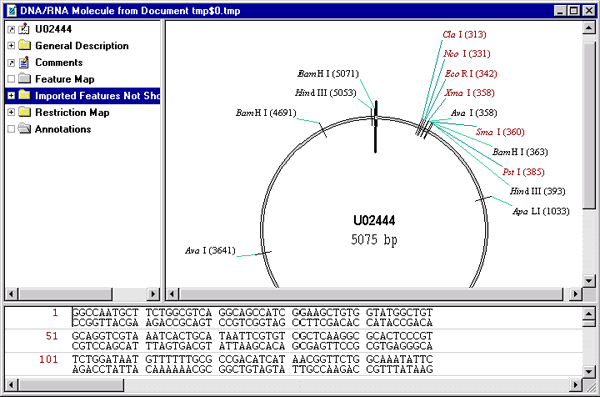
5. Display the result of the query using MIME type association
The alternative way to transfer search results from the NCBI server to Vector NTI is to use MIME type associations of your browser. Vector NTI automatically configures itself as a helper application for Microsoft Internet Explorer and Netscape Communicator. If you have Netscape Navigator v.3.x or earlier or some other browser, skip this step or see the corresponding part of the User Manual for configuration instructions.
Switch back to the U02444 / pKK388-1GenBank report and scroll it to the “Save” section at the bottom. Choose the PC (or Macintosh) / MIME format and press the Save button. At this moment your browser will try to load the document into the associated application. If Vector NTI was configured correctly, a new window should appear in its workspace showing the U02444 molecule.
6. Use Alignment and Analysis tools
The standard distribution of Vector NTI has several tools to perform sequence alignment and analysis. To perform the alignment, switch to the Database Explorer window (or open it using the Local Database button on the main toolbar of the Vector NTI workspace) and select the Proteins table from the combo box.
Vector NTI displays the list of proteins in the database. Shift-click on 41BB_HUMAN and 41BB_MOUSE to select them and choose the Align 4
Multiple Sequences on BCM Server command from the Database Explorer’s Tools menu.
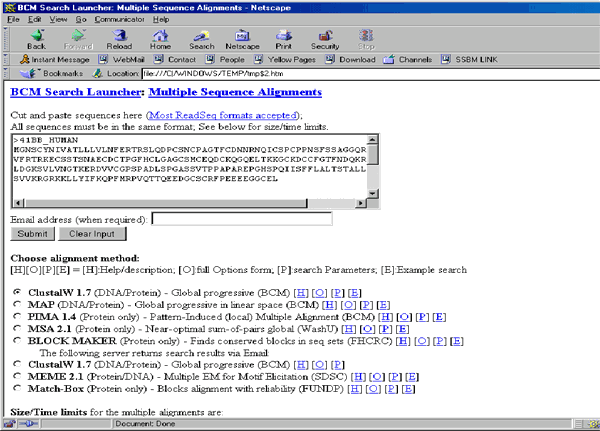
Vector NTI invokes the browser and displays the BCM Search page containing the sequences of the selected proteins in FASTA format. Press the Submit button to start the ClustalW alignment (the default option). If the BCM server is not too busy, you will receive the response in 5-10 seconds.
Now let’s try to analyze a protein using the ProtScale program on the ExPASy server in Switzerland. Switch back to the Database Explorer window and click on the 41_HUMAN molecule to select it. Then, choose the “Protein Scales (hydrophobicity etc.) via ProtScale on ExPASy Server” command from the Database Explorer’s Analyze menu.
Again, Vector NTI invokes the browser and displays the ProtScale page containing the sequence of 41_HUMAN. Scroll to the bottom of the page and press the Submit button to start the default analysis (hydrophobicity / Kyte & Doolitle). If the ExPASy server is not too busy, you will receive the response in 2-5 seconds. Scroll down the results page to see results in numerical and graphical form.
6. Saving search results as comments
Vector NTI allows you to associate any text data with database objects in the form of comments. Let’s try to perform a simple PROSITE database search and save the results to the database for future reference.
Activate the Database Explorer window and click on the 41_HUMAN protein to select it. Choose the “Compare Against 4
PROSITE Database via ScanProsite on ExPASy Server” command from the Database Explorer’s Tools menu.
When you see the ScanProsite page in the browser, press the “Start The Scan” button. When the results are ready, select the resulting hits by dragging the mouse across the page and choose the Copy command from your browser’s Edit menu to put results on the Clipboard (don’t select the sequence; it is already stored in the Vector NTI database).
Now switch back to the Database Explorer window (41_HUMAN should still be selected) and choose the Edit command from the Explorer’s Protein menu.
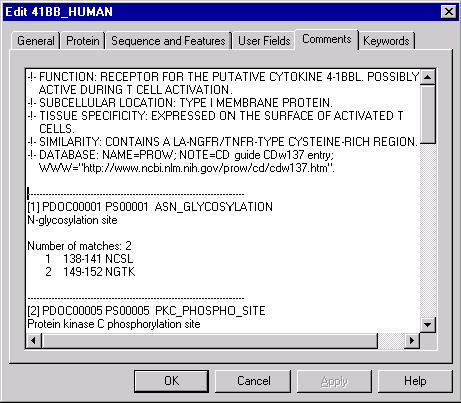
Vector NTI displays a tabbed dialog allowing you to edit various information associated with the protein molecule. Click on the Comments tab, scroll down the comments text and click to set the text cursor for insertion. Now press control-V (command-V on Macintosh) to paste the search results from the Clipboard. Edit the resulting text to your liking and press the OK button to save it to the database.
The search results are now saved in the database and you can see them when necessary. In addition to the Edit dialog, you can view the comments from the 41_HUMAN’s Display window by double-clicking on the Comments line in the text pane.
7. Close the Molecule Display windows and exit Vector NTI
You have now taken a first look at Vector NTI’s Tools and Internet connection. Close all Display windows using the Close command in the Molecule menu, and then quit the program with the File menu’s Exit command (Windows) or Quit command (Macintosh). If you are not going to continue your work with the Internet, you can close the WWW browser and disconnect, if required.
See Also:
Chapter 14 -- Molecule Components and Construction
Chapter 17 -- Vector NTI Tools
Chapter 7 -- Molecule Construction
Introduction
Chapter 2 -- Molecule Operations
Chapter 5 -- Database Explorer
Chapter 11 -- Display Windows and User Interface
Chapter 15 -- Database Operations
Chapter 12 -- Gel Display Windows
Chapter 13 -- Oligonucleotides and Primers
Back to Index