- Background
- Your Task: Complex cloning
- First Design: Complicated Recipient
- Second Design: Complicated Donor
Note This chapter assumes that you have completed the preceding tutorials and are familiar with Vector NTI’s user interface, selection of molecule fragments, the Goal List and molecule design with the aid of Vector NTI’s built-in biological knowledge.
This chapter introduces you to more complex Vector NTI cloning situations. Now that you have completed a simple molecule design, let’s look at two more complicated cases. First we will assign some complex conditions to the recipient molecule while leaving the donor relatively simple; then, we will use a relatively simple recipient but make the donor more complex. Since you are probably getting tired of pBR322 and pUC19, let’s change molecules. We will now use BPV1 and SV40, and our overall task will be to clone the large T-cell antigen gene (LARGE_T) from SV40 into BPV1.
Your Task: Complex cloning
Your task will be to clone the LARGE_T gene from SV40 into BPV1 in two different ways. You will place requirements first upon the recipient and then upon the donor, and consider Vector NTI’s proposed design plans.
Follow the steps in the order shown. Figures show what your screen should look like at various points along the way.
1. Launch Vector NTI
Launch Vector NTI by double-clicking its icon in the program group or folder in which you installed Vector NTI.
2. Open and Arrange Display windows for BPV1 and SV40
Open Molecule Display windows containing the molecules BPV1 and SV40. Use the default Display settings. You will be working with the molecules’ graphical maps, so arrange the Display windows conveniently.
First Design: Complicated Recipient
In the first complex design, you will insert SV40’s LARGE_T gene into the second ApaLI site of BPV1. You will also require that Vector NTI save the 5’ ApaLI site and prohibit blunt–blunt fragments. If the donor has ApaLI sites appropriate for cutting the cloned fragment, then the problem will be simple. If not, the system will have to take a more complicated approach to perform the insertion.
3. Define the recipient
Switch to the BPV1 Display window and click somewhere in its graphics pane to activate it. Call up the Fragment Wizard, select the Design Recipient option on the first screen and press the Next button. To define 5’ terminus, click on the label of the ApaLI restriction site #2 in the graphics pane (nucleotide 7631, or about the 11 o’clock position). Click the Next button to continue. The next screen allows you to specify the save/lose condition for ApaLI site on 5’ end of the recipient. Select Save Site and press the Next button to go to the 3’ terminus screen. Now hold down the shift key and click on the same site again. The name and the position of the site are displayed in the Fragment Wizard. Press the Finish button to complete the definition of the recipient fragment.
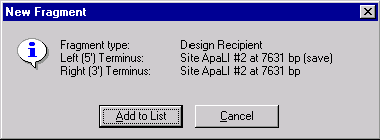
The New Fragment message box is displayed with the description of the selected recipient fragment. Check the description of the fragment in the message box. Notice, that the 3’ and 5’ termini of the recipient fragment are set on the same ApaLI site. Press the Add to List button. The recipient fragment is added to the Molecule Goal list.
4. Define the donor
Switch to the SV40 Display window and click somewhere in its graphics pane to activate it. Call up the Fragment Wizard, select the Design Donor option on the first screen and press the Next button. Click on the LARGE_T signal’s symbol or label in the graphics pane to select it and press the Finish button in the Fragment wizard.
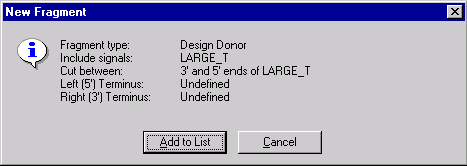
Check the description of the donor fragment in the message box and press Add to List button. The donor fragment is added to the Molecule Goal List.
5. Inspect the Goal Molecule Definition List
Now check the Goal Molecule Definition List. Press the Open Goal List button (  ) in the main toolbar and the Construct/Design Molecule dialog box appears. The Component Fragments section of the dialog box contains the Goal Molecule Definition List. The list consists of the two fragments you defined. The recipient fragment must be the first in the Goal Molecule Definition List.
) in the main toolbar and the Construct/Design Molecule dialog box appears. The Component Fragments section of the dialog box contains the Goal Molecule Definition List. The list consists of the two fragments you defined. The recipient fragment must be the first in the Goal Molecule Definition List.
Double-click the SV40 fragment, and the Fragment Editor dialog box opens. Click the "Inverted” check box to change LARGE_T’s direction to match the recipient’s direction and press OK. (You could leave LARGE_T in its original orientation if you want to; the system will design your new molecule either way. We have changed LARGE_T to inverted only to demonstrate how Vector NTI can clone fragments in different orientations.)
6. Enter general information for your new molecule
Change the molecule’s name to TUTORIAL3. Press the General Info button. In the Description field, enter “Tutorial molecule #3”. Set the Replicon Type to Plasmid, and turn on the Bacteria Extra-Chromosome Replication option. Enter your name as a keyword. Press OK to return to the Construct/Design dialog box. Check the Recipient’s Start button in the radio button group just above the Component Fragments section to ask Vector NTI to make the start of the new molecule at the same place (if possible) where the recipient molecule (BPV1) starts.
7. Prepare to design the new molecule
Press the Design button in the upper right corner of the Construct/Design Molecule dialog box. Vector NTI first asks you to name a subbase for results. Select the “Tutorial” subbase we created before and press OK to continue. Vector NTI saves the information you have entered about your new molecule, i.e. the name, replicon type, extra-chromosome replication abilities, list of fragments, etc.
The Design Parameters dialog box appears. Leave all the settings at their default values and move on to the next step.
8. Set the design preferences
Now click on the Preferences button. The Design Preferences dialog box appears. Note that the blunt–blunt ligation box is already turned off. This is because Vector NTI remembers the changes you made in Chapter 7 of the Tutorial. The program remembers your design preferences so that you won’t have to set them every time you design a new molecule.
Below the check boxes are priority lists specifying which of the techniques are preferable. Leave these at their default values.
Press the OK button to accept the Design Preferences and return to the Design Parameters dialog box.
9. Design the new molecule
With the goal molecule defined and your preferences for design techniques entered and prioritized, you are ready to command Vector NTI to design your new molecule.
Press the Start Design button. Vector NTI gathers information from the database, generates some necessary data and then begins designing your goal molecule. “Thinking...” appears in an information box. As before, an optimum cloning method is found within a few minutes and the goal molecule is constructed based on that best option.
10. Inspect the new molecule
When the design is complete Vector NTI opens a new Molecule Display window containing the molecule you have created.
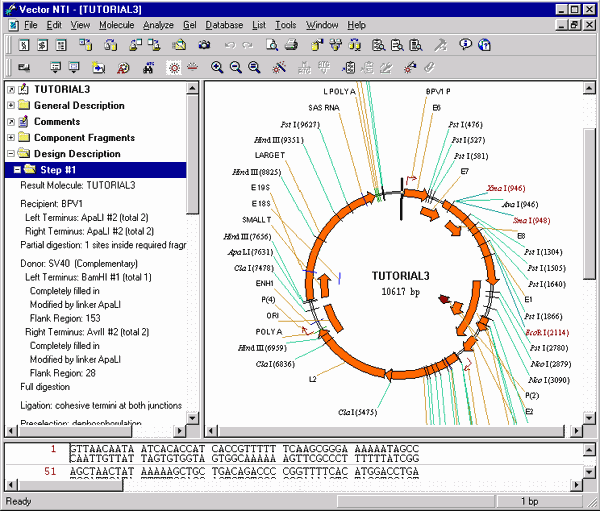
11. Inspect the graphical map and text description of your new molecule
Switch to the text pane and open TUTORIAL3’s Design Description folder and the Step #1 subfolder. A description of how to create TUTORIAL3 appears.
Design Description
Step #1
Result Molecule: TUTORIAL3
Recipient: BPV1
Left Terminus: ApaLI #2 (total 2)
Right Terminus: ApaLI #2 (total 2)
Partial digestion: 1 sites inside required fragment, 2 sites total
Donor: SV40 (Complementary)
Left Terminus: BamHI #1 (total 1)
Completely filled in
Modified by linker ApaLI
Flank Region: 153
Right Terminus: AvrII #2 (total 2)
Completely filled in
Modified by linker ApaLI
Flank Region: 28
Full digestion
Ligation: cohesive termini at both junctions
Preselection: dephosphorylation
Transformation system: Bacteria
Extra-chromosomal replication
Recommended method(s) for clone analysis:
Restriction Analysis
Recommended enzyme(s) for restriction analysis:
AatI
Recommended enzyme(s) for restriction analysis
defining clone with required fragment orientation:
AvrII
ApaLI
Recommended oligonucleotide for colony hybridization
of cloned fragment in resulting molecule:
CTAGGCTTTTGCAAAAAGCT
Recommended primers for PCR amplification
of cloned fragment in resulting molecule:
Sense Primer:
ATGTCTGTACCGCCATCGGTGCAC
Antisense Primer:
ACCTATATCGGTGCACGGGG
Important Restriction Sites:
Cloned fragment may be cut by AvrII and ApaLI
Recombinant lacks sites of:
ApaI AscI BsiWI Bsp120I BssHII Ecl136II
Eco47III EcoRV KspI MluNI NaeI NgoMI
NotI PacI PaeR7I PmeI PvuI SacI
SalI SnaBI
Unique sites outside cloned fragment:
AatII Acc65I AccIII AflII AosI AsuII EagI EcoRI
KpnI MluI NruI Psp1406I SmaI XbaI XmaI
Recall that we defined a specific ApaLI site on the recipient molecule as the site into which the donor fragment was to be cloned and that we required Vector NTI to save this site in the recombinant. ApaLI was not a unique site, so Vector NTI had to use partial digestion to isolate the recipient. Then the system had to find a way to insert the donor fragment. There may have been no good sites available on the donor to fit into the recipient’s ApaLI site; so, Vector NTI cut out the donor fragment with convenient sites which did not require partial digestion and left short flank regions (BamHI and AvrII). These termini were then filled in completely and ApaLI linkers were attached to the blunt ends. This fragment was then cloned into the recipient.
You can be sure the cloned fragment contains no ApaLI sites (Vector NTI checked this when selecting a linker) and that there were no easier ways to perform this cloning.
The rest of TUTORIAL3’s Design Description folder is by and large similar to the Design Description of TUTORIAL2, which we designed in Part 3 of the Tutorial. However, because the termini of the recipient are identical for TUTORIAL3, we need to know how to distinguish clones with the proper orientation. Thus a new paragraph appears in TUTORIAL3’s design description, recommending that the AvrII and ApaLI restriction sites would allow you to distinguish correct and parasitic orientations in gel.
You may print out this design plan if you wish, and then proceed to the next design task.
Second Design: Complicated Donor
We will now design a second molecule based on BPV1 and SV40; this time we will make the donor more complicated rather than the recipient.
12. Define recipient
Switch to the BPV1 Display window’s graphics pane and call up the Fragment Wizard. On the first screen of the Fragment screen select the Design Recipient option and press the Next button. In the second screen, select the “Set to a position” option and enter 5000 as the position of a 5’ terminus. Press the Next button to go to the 3’ terminus screen. Select the “Set to a position” option again and enter 2500. Press the Finish button, inspect the results in a message box and press the Add to List button. The recipient fragment is added to the Molecule Goal List.
13. Define donor
Switch to the SV40 Display window’s graphics pane and call up the Fragment Wizard. On the first screen of the Fragment screen select the Design Donor option and press the Next button. Click on the LARGE_T signal’s symbol or label in the graphics pane to select it and press the Next button in the Fragment wizard. The default option on the next screen is “Leave terminus Undefined”. Do not change this option; press the Next button to go to the flank region screen.
Here you can specify the maximum size of the flank region or let Vector NTI use all available space outside the selected signal. Select the “Use flank region no larger than” option. Fragment Wizard prompts you that you can either enter the maximum length in the edit box or select it in the graphics pane. Move your cursor over the 5’ end of the selection wireframe in the graphics pane (the cursor shape should change to a crosshair with 5’ symbol), press and hold down the mouse button to grab the 5’ end of the wireframe and drag it to the position about nucleotide 2250 (current position is displayed in the status bar). The edit box in the Fragment wizard shows you the maximum size of the flank region (it should be about 400 nucleotides). When you are finished, press the Next button to go to the 3’ terminus screen.
Let’s specify here that Vector NTI should use the NcoI restriction site at nucleotide 38 to cut the 3’ terminus of the donor fragment. Select the “Use specific site” option, press and hold down the shift key and click on the NcoI restriction site at nucleotide 38 (scroll the graphics pane down if needed). The name and the position of the restriction site appears in the Fragment wizard screen. Now press the Finish button to complete the definition of the donor fragment.
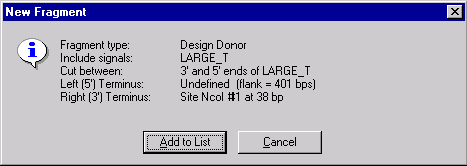
The New Fragment message box is displayed with the description of the selected donor fragment. Check the description of the fragment in the message box. Note that the 5’ terminus is described as Undefined with the specified maximum length of a flank region and the 3’ terminus is set to the NcoI site. Press the Add to List button. The donor fragment is added to the Molecule Goal list.
14. Inspect the Goal List
Call up the Construct/Design Molecule dialog box and inspect the fragments you have defined. The recipient fragment must be the first in the Goal Molecule Definition List. Double-click the donor fragment, the Fragment Editor appears. Note that one end of the donor is determined by a restriction site, while the other end has a flank region defined. This makes the situation considerably more complicated than our previous cases, where the donors were more simply defined. Make the donor inverted and press OK.
15. Design the new molecule
Enter our usual information about the new molecule.
Change the name to TUTORIAL4 and set the other options as usual. Check the Recipient’s Start button in the radio button group just above the Component Fragments section to ask Vector NTI to make the start of the new molecule at the same place (if possible) where the recipient molecule (BPV1) starts.
Press the Design button in the upper right corner of the Construct/Design Molecule dialog box. Vector NTI first asks you to name a subbase for results; select the “Tutorial” subbase and press OK to continue. Leave the Design Preferences at their default settings and press the Start Design button. Vector NTI thinks for a while and then produces a workable design for the molecule you have requested.
16. Inspect and print the new molecule
Open a new Molecule Display window containing TUTORIAL4. Inspect the Step #1 subfolder of the Design Description folder. Notice how in this case, despite the more complex conditions we had required, Vector NTI found a simple and convenient design. Both donor and recipient can be isolated at the NcoI and BamHI sites, so the cloning is straightforward.
We had specified a maximum flank region of about 400 nucleotides; Vector NTI found a good BamHI site, allowing the actual flank region to stay within the assigned limit.
If you wish, print out the design description and/or graphical map of TUTORIAL4.
See Also:
Chapter 8 -- Molecule Design
Introduction
Chapter 7 -- Molecule Construction
Chapter 14 -- Molecule Components and Construction
Chapter 15 -- Database Operations
Chapter 16 -- Molecule Design
Chapter 11 -- Display Windows and User Interface
Chapter 13 -- Oligonucleotides and Primers
Chapter 18 -- Reports Generation
Back to Index